8 Introduction to Python and Pandas
This session is typically ran held in parallel to the Introduction to R and Tidyverse. Participants of the summer schools chose which to attend based on their prior experience. We recommend the introduction to R session if you have no experience with neither R nor Python.
For this chapter’s exercises, if not already performed, you will need to download the chapter’s dataset, decompress the archive, and create and activate the conda environment.
Do this, use wget or right click and save to download this Zenodo archive: 10.5281/zenodo.11394586, and unpack
tar xvf python-pandas.tar.gz
cd python-pandas/You can then create the subsequently activate environment with
conda env create -f python-pandas.yml
conda activate python-pandasOver the last few years, Python has gained popularity thanks to the numerous libraries (packages with pre-written functions) in bioinformatics, statistical data analysis, and machine learning. While a few years ago, it was often necessary to go to R for performing routine data manipulation and analysis tasks, nowadays Python has a vast ecosystem of useful libraries for working on metagenomic data. Existing libraries exist for many different file formats encountered in metagenomics, such as fasta, fastq, sam, bam, etc. Furthermore, python is fast and extremely useful for writing programs that can be easily called from the command line like many existing tools.
This tutorial/walkthrough will provide a short introduction to the popular libraries for data analysis pandas ((https://pandas.pydata.org/). This library has functions for reading and manipulating tabular data similar to the data.frame() in R together with some basic data plotting. This will set the base for learning Python and use it for data analysis.
There are many IDEs in which Python code can be written. For data analysis, Jupyter is powerful and popular which looks and functions similar to R markdown, where code is written in code blocks with space in text blocks for annotations. In this tutorial/walkthrough, we will use these notebooks for running and visualising Python code.
Learning objectives:
- Get familiar with the Python code syntax and use Jupyter Notebook for executing code
- Get a kickstart to utilising the endless possibilities of data analysis in Python that can be applied to our data
8.1 Working in a Jupyter environment
This tutorial/walkthrough is using a Jupyter Notebook (https://jupyter.org) for writing and executing Python code and for annotating.
Jupyter notebooks have two types of cells: Markdown and Code. The Markdown cell syntax is very similar to R markdown. The markdown cells are used for annotating code, which is important for sharing work with collaborators, reproducibility, and documentation.
Change the directory to the the working directory of this tutorial/walkthrough.
cd python-pandas_lecture/To launch jupyter, run the following command in the terminal. This will open a browser window with jupyter running.
jupyter notebookJupyter Notebook should have a file structure with all the files from the working directory. Open the student-notebook.ipynb notebook by clicking on it. This notebook has exactly the same code as written in this book chapter and is only a support so that it is not necessary to copy and paste the code. It is of course also possible to copy the code from this chapter into a fresh notebook file by clicking on: File > New > Notebook.
If you cannot find student-notebook.ipynb, it is possible the working directory is not correct. Make sure that pwd returns /<path>/<to>/python-pandas/python-pandas_lecture.
8.1.1 Creating and running cells
There are multiple ways of making a new cells in jupyter, such as typing the letter b, or using the cursor on the bottom of the page that says click to add cell. The cells can be assigned to code or markdown using the drop down menu at the top. Code cells are always in edit mode. Code can be run with pressing Shift + Enter or click on the ▶ botton. To make an markdown cell active, double-click on a markdown cell, it switches from display mode to edit mode. To leave the editing mode by running the cell.
Before starting it might be nice to clear the output of all code cells, by clicking on:
edit > Clear outputs of All Cells
8.1.2 Markdown cell syntax
Here a few examples of the syntax for the Markdown cells are shown, such as making words bold, or italics. For a more comprehensive list with syntax check out this Jupyter Notebook cheat-sheet (https://www.ibm.com/docs/en/watson-studio-local/1.2.3?topic=notebooks-markdown-jupyter-cheatsheet).
List of markdown cell examples:
**bold**: bold_italics_: italics
Code
- `inline code` :
inline code
LaTeX maths
$ x = \frac{\pi}{42} $: \[ x = \frac{\pi}{42} \]
URL links
[link](https://www.python.org/): link
Images

In many cases, there are multiple syntaxes, or ‘ways of doing things,’ that will give the same results. For each section in this tutorial/walkthrough, one way is presented.
8.1.3 code cell syntax
The code cells can interpret many different coding languages including Python and Bash. The syntax of the code cells is the same as the syntax of the coding languages, in our case python.
Below are some examples of Python code cells with some useful basic python functions:
print() is a python function for printing lines in the terminal
print() is the same as echo in bash
print("Hello World from Python!")Hello World from Python!It is also possible to run bash commands in Jupyter, by adding a ! at the start of the line.
! echo "Hello World from bash!"Hello World from bash!Stings or numbers can be stored as a variable by using the = sign.
i = 0Ones a variable is set in one code cell they are stored and can be accessed in other downstream code cells.
To see what value a variable contains, the print() function can be used.
print(i)0You can also print multiple things together in one print statement such as a number and a string.
print("The number is", i, "Wow!")The number is, 0, Wow!8.2 Pandas
8.2.1 Getting started
Pandas is a Python library used for data manipulation and analysis.
We can import the library like this.
import pandas as pdWe set pandas to the alias pd because we are lazy and do not want to write the full word too many times.
Now that Pandas is imported, we can check if it worked correctly, and check which version is running by runing .__version__.
pd.__version__‘2.2.2’
8.2.2 Pandas data structures
The primary data structures in Pandas are the Series and the DataFrame. A Series is a one-dimensional array-like object containing a value of the same type and can be imagined as one column in a table Figure 8.1. Each element in the series is associated with an index from 0 to the number of elements, but these can be changed to labels. A DataFrame is two-dimensional, and can change in size after it is created by adding and removing rows and columns, which can hold different types of data such as numbers and strings Figure 8.2. The columns and rows are labelled. By default, rows are unnamed and are indexed similarly to a series.

Series. The dark grey squares are the index or row names, and the light grey squares are the elements.

For a more in detail pandas getting started tutorial click here (https://pandas.pydata.org/docs/getting_started/index.html#)
8.3 Reading data with Pandas
Pandas can read in csv (comma separated values) files, which are tables in text format. It is called _c_sv because each value is separated from the others through a comma.
A,B
5,6
8,4Another common tabular separator are tsv, where each value is separated by a tab \t.
A\tB
5\t6
8\t4The dataset that is used in this tutorial/walkthrough is called "all_data.tsv", and is tab-separated. Pandas by default assume that the file is comma delimited, but this can be change by using the sep= argument.
pd.read_csv() is the pandas function to read in tabular tables. The sep= can be specified argument, sep=, is the default.
df = pd.read_csv("../all_data.tsv", sep="\t")
df| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 | 3 | 11 |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | 11 |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 | 3 | 11 |
| 3 | 6182 | 1984 | Graduation | Together | 26646 | 1 | 0 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | 0 | 4 | 6 | 0 | 3 | 11 |
| 4 | 7446 | 1967 | Master | Together | 62513 | 0 | 1 | 520 | 42 | 98 | 0 | 42 | 14 | 6 | 4 | 10 | 6 | 0 | 3 | 11 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1749 | 9432 | 1977 | Graduation | Together | 666666 | 1 | 0 | 9 | 14 | 18 | 8 | 1 | 12 | 3 | 1 | 3 | 6 | 0 | 3 | 11 |
| 1750 | 8372 | 1974 | Graduation | Married | 34421 | 1 | 0 | 3 | 3 | 7 | 6 | 2 | 9 | 1 | 0 | 2 | 7 | 0 | 3 | 11 |
| 1751 | 10870 | 1967 | Graduation | Married | 61223 | 0 | 1 | 709 | 43 | 182 | 42 | 118 | 247 | 9 | 3 | 4 | 5 | 0 | 3 | 11 |
| 1752 | 7270 | 1981 | Graduation | Divorced | 56981 | 0 | 0 | 908 | 48 | 217 | 32 | 12 | 24 | 2 | 3 | 13 | 6 | 0 | 3 | 11 |
| 1753 | 8235 | 1956 | Master | Together | 69245 | 0 | 1 | 428 | 30 | 214 | 80 | 30 | 61 | 6 | 5 | 10 | 3 | 0 | 3 | 11 |
When you are unsure what arguments a function can take, it is possible to get a help documentation using help(pd.read_csv)
In most cases, data will be read in with the pd.read_csv() function, however, internal Python data structures can also be transformed into a pandas data frame. For example using a nested list, were each row in the datafram is a list [].
df = pd.DataFrame([[5,6], [8,4]], columns=["A", "B"])
df| A | B | |
|---|---|---|
| 0 | 5 | 6 |
| 1 | 8 | 4 |
Another usful transformation is from a dictionary (https://docs.python.org/3/tutorial/datastructures.html#dictionaries) to pd.Dataframe.
table_data = {'A' : [5, 6]
'B' : [8, 4]}
df = pd.DataFrame(table_data)
df| A | B | |
|---|---|---|
| 0 | 5 | 6 |
| 1 | 8 | 4 |
There are many ways to turn a DataFrame back into a dictonary (https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_dict.html#pandas.DataFrame.to_dict), which might be very handy for certain purposes.
8.4 Data exploration
The data for this tutorial/walkthrough is from a customer personality analysis of a company trying to better understand how to modify their product catalogue. Here is the link to the original source (https://www.kaggle.com/datasets/imakash3011/customer-personality-analysis) for more information.
8.4.1 Columns
To display all the column names from the imported DataFrame, the attribute columns can be called.
df.columnsIndex(['ID', 'Year_Birth', 'Education', 'Marital_Status', 'Income', 'Kidhome',
'Teenhome', 'MntWines', 'MntFruits', 'MntMeatProducts',
'MntFishProducts', 'MntSweetProducts', 'MntGoldProds',
'NumWebPurchases', 'NumCatalogPurchases', 'NumStorePurchases',
'NumWebVisitsMonth', 'Complain', 'Z_CostContact', 'Z_Revenue'],
dtype='object')Each column has its own data types which are highly optimised. A column with only integers has the data type int64. Columns with decimal numbers are called float64. A column with only strings, or a combination of strings and integers or floats is called an object.
df.dtypesID int64
Year_Birth int64
Education object
Marital_Status object
Income float64
Kidhome int64
Teenhome int64
MntWines int64
MntFruits int64
MntMeatProducts int64
MntFishProducts int64
MntSweetProducts int64
MntGoldProds int64
NumWebPurchases int64
NumCatalogPurchases int64
NumStorePurchases int64
NumWebVisitsMonth int64
Complain int64
Z_CostContact int64
Z_Revenue int64
dtype: objectThe 64 indicates the number of bits the integers are stored in. 64 bits is the largest pandas handels. When it is known that a value is in a certain range, it is possible to change the bits to 8, 16, or 32. This chosing the correct range might reduce memory usage, to be save, 64 range is so large it will incorporate most user cases.
df['Kidhome'] = df['Kidhome'].astype('int8')8.4.2 Inspecting the DataFrame
To quickly check how many rows and columns the DataFrame has, we can access the shape attribute.
df.shape(1754, 20)The .shape attribute of a DataFrame provides a tuple representing its dimensions. A tuple is a Python data structure that is used to store ordered items. In the case of shape, the first item is always the row, and the second item is the columns. To print, or access the rows or columns the index can be used. .shape[0] gives the number of rows, and .shape[1] gives the number of columns.
df.shape[0]1754df.shape[1]20It is often useful to have a quick look at the first rows, to get, for example, an idea of the data was read correctly. This can be done with the head() function.
df.head()| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 | 3 | 11 |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | 11 |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 | 3 | 11 |
| 3 | 6182 | 1984 | Graduation | Together | 26646 | 1 | 0 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | 0 | 4 | 6 | 0 | 3 | 11 |
| 4 | 7446 | 1967 | Master | Together | 62513 | 0 | 1 | 520 | 42 | 98 | 0 | 42 | 14 | 6 | 4 | 10 | 6 | 0 | 3 | 11 |
The difference between calling a function and an atribute is the (). .head() is a function and will perform an action. While .shape is an attribute and will return a value that is already stored in the DataFrame.
What we can see it that, unlike R, Python and in extension Pandas is 0-indexed instead of 1-indexed.
8.4.3 Accessing rows and columns
It is possible to access parts of the data in DataFrames in different ways. The first method is sub-setting rows using the row name and column name. This can be done with the .loc, which loc ates row(s) by providing the row name and column name [row, column]. When the rows are not named, the row index can be used instead. To print the second row, this would be index 1 since the index in Python starts at 0. To print the all the columns, the : is used.
df.loc[1, :]ID 2174
Year_Birth 1954
Education Graduation
Marital_Status Single
Income 46344.0
Kidhome 1
Teenhome 1
MntWines 11
MntFruits 1
MntMeatProducts 6
MntFishProducts 2
MntSweetProducts 1
MntGoldProds 6
NumWebPurchases 1
NumCatalogPurchases 1
NumStorePurchases 2
NumWebVisitsMonth 5
Complain 0
Z_CostContact 3
Z_Revenue 11
Name: 1, dtype: objectTo print a range of rows, the first and last index can be written with a :. To print the second and third row, this would be [1:2, :].
df.loc[1:2]| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | 11 |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 | 3 | 11 |
To print all the rows with a certain column name works the same as subsetting rows, but then adding the column name after the comma.
df.loc[:, "Year_Birth"]0 1957
1 1954
2 1965
3 1984
4 1967
...
1749 1977
1750 1974
1751 1967
1752 1981
1753 1956It is important to notice that almost all operations on DataFrames are not in place, meaning that the DataFrame is not modified. To keep the changes, the DataFrame has to be actively stored using the same name or a new variable.
To save the changes, a new DataFrame has to be created, or the existing DataFrame has to be overwritten. This can be done by directing the output to a variable with =. To make a new DataFrame with only the “Education” and “Marital_Status” columns, the column names have to be placed in a list ['colname1', 'colname2'].
df.head()
new_df = df.loc[:, ["Education", "Marital_Status"]]
new_df| Education | Marital_Status | |
|---|---|---|
| 0 | Graduation | Single |
| 1 | Graduation | Single |
| 2 | Graduation | Together |
| 3 | Graduation | Together |
| 4 | Master | Together |
| … | … | … |
| 1749 | Graduation | Together |
| 1750 | Graduation | Married |
| 1751 | Graduation | Married |
| 1752 | Graduation | Divorced |
| 1753 | Master | Together |
| 1754 rows × 2 columns |
It is also possible to remove rows and columns from a DataFrame. This can be done with the function drop(). To remove the columns Z_CostContact and Z_Revenue and keep those changes, it is necessary to overwrite the DataFrame. To make sure Pandas understands that the its columns that need to be removed, the axis can be specified. Rows are called axis=0, and columns are called axis=1. In most cases Pandas will guess correctly without specifying the axis, since in this case the no row is called Z_CostContact or Z_Revenue. It is however good practice to add the axis to make sure Pandas is operating as expected.
df = df.drop("Z_CostContact", axis=1)
df = df.drop("Z_Revenue", axis=1)df = df.drop(["Z_CostContact", "Z_Revenue"], axis=1)8.4.4 Conditional subsetting
So far, all the subsetting has been based on row names and column names. However, in many cases, it is more helpful to look only at data that contain certain items. This can be done using conditional subsetting, which is based on Boolean values True or False. pandas will interpret a series of True and False values by printing only the rows or columns where a True is present and ignoring all rows or columns with a False.
For example, if we are only interested in individuals in the table who graduated, we can test each string in the column Education to see if it is equal (==) to Graduation. This will return a series with Boolean values True or False.
education_is_grad = (df["Education"] == "Graduation")
education_is_grad0 True
1 True
2 True
3 True
4 False
...
1749 True
1750 True
1751 True
1752 True
1753 False
Name: Education, Length: 1754, dtype: boolTo quicky check if the True and False values are correct, it can be useful to print out this column.
df["Education"]0 Graduation
1 Graduation
2 Graduation
3 Graduation
4 Master
… …
1749 Graduation
1750 Graduation
1751 Graduation
1752 Graduation
1753 Master
Name: Education, length: 1754, dtype: objectIt is possible to provide pandas with multiple conditions at the same time. This can be done by combining multiple statements with &.
two_at_once = (df["Education"] == "Graduation") & (df["Marital_Status"] == "Single")
two_at_once0 True
1 True
2 False
3 False
4 False
...
1749 False
1750 False
1751 False
1752 False
1753 False
Length: 1754, dtype: boolTo find out the total number of Graduated singles, the .sum() can be used.
sum(two_at_once)252These Series of Booleans can be used to subset the dataframe to rows where the condition(s) are True:
df[two_at_once]| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 | 3 | 11 |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | 11 |
| 18 | 7892 | 1969 | Graduation | Single | 18589 | 0 | 0 | 6 | 4 | 25 | 15 | 12 | 13 | 2 | 1 | 3 | 7 | 0 | 3 | 11 |
| 20 | 5255 | 1986 | Graduation | Single | NaN | 1 | 0 | 5 | 1 | 3 | 3 | 263 | 362 | 27 | 0 | 0 | 1 | 0 | 3 | 11 |
| 33 | 1371 | 1976 | Graduation | Single | 79941 | 0 | 0 | 123 | 164 | 266 | 227 | 30 | 174 | 2 | 4 | 9 | 1 | 0 | 3 | 11 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1720 | 10968 | 1969 | Graduation | Single | 57731 | 0 | 1 | 266 | 21 | 300 | 65 | 8 | 44 | 8 | 8 | 6 | 6 | 0 | 3 | 11 |
| 1723 | 5959 | 1968 | Graduation | Single | 35893 | 1 | 1 | 158 | 0 | 23 | 0 | 0 | 18 | 3 | 1 | 5 | 8 | 0 | 3 | 11 |
| 1743 | 4201 | 1962 | Graduation | Single | 57967 | 0 | 1 | 229 | 7 | 137 | 4 | 0 | 91 | 4 | 2 | 8 | 5 | 0 | 3 | 11 |
| 1746 | 7004 | 1984 | Graduation | Single | 11012 | 1 | 0 | 24 | 3 | 26 | 7 | 1 | 23 | 3 | 1 | 2 | 9 | 0 | 3 | 11 |
| 1748 | 8080 | 1986 | Graduation | Single | 26816 | 0 | 0 | 5 | 1 | 6 | 3 | 4 | 3 | 0 | 0 | 3 | 4 | 0 | 3 | 11 |
252 rows × 20 columns
It is not actually necessary to create a series every time for subsetting the table and it can be done in one go by combining the conditions within the df[].
df[(df["Education"] == "Master") & (df["Marital_Status"] == "Single")]| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26 | 10738 | 1951 | Master | Single | 49389 | 1 | 1 | 40 | 0 | 19 | 2 | 1 | 3 | 2 | 0 | 3 | 7 | 0 | 3 | 11 |
| 46 | 6853 | 1982 | Master | Single | 75777 | 0 | 0 | 712 | 26 | 538 | 69 | 13 | 80 | 3 | 6 | 11 | 1 | 0 | 3 | 11 |
| 76 | 11178 | 1972 | Master | Single | 42394 | 1 | 0 | 15 | 2 | 10 | 0 | 1 | 4 | 1 | 0 | 3 | 7 | 0 | 3 | 11 |
| 98 | 6205 | 1967 | Master | Single | 32557 | 1 | 0 | 34 | 3 | 29 | 0 | 4 | 10 | 2 | 1 | 3 | 5 | 0 | 3 | 11 |
| 110 | 821 | 1992 | Master | Single | 92859 | 0 | 0 | 962 | 61 | 921 | 52 | 61 | 20 | 5 | 4 | 12 | 2 | 0 | 3 | 11 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1690 | 3520 | 1990 | Master | Single | 91172 | 0 | 0 | 162 | 28 | 818 | 0 | 28 | 56 | 4 | 3 | 7 | 3 | 0 | 3 | 11 |
| 1709 | 4418 | 1983 | Master | Single | 89616 | 0 | 0 | 671 | 47 | 655 | 145 | 111 | 15 | 7 | 5 | 12 | 2 | 0 | 3 | 11 |
| 1714 | 2980 | 1952 | Master | Single | 8820 | 1 | 1 | 12 | 0 | 13 | 4 | 2 | 4 | 3 | 0 | 3 | 8 | 0 | 3 | 11 |
| 1738 | 7366 | 1982 | Master | Single | 75777 | 0 | 0 | 712 | 26 | 538 | 69 | 13 | 80 | 3 | 6 | 11 | 1 | 0 | 3 | 11 |
| 1747 | 9817 | 1970 | Master | Single | 44802 | 0 | 0 | 853 | 10 | 143 | 13 | 10 | 20 | 9 | 4 | 12 | 8 | 0 | 3 | 11 |
75 rows × 20 columns
8.4.5 Describing a DataFrame
Sometimes is is nice to get a quick overview of the data in a table, such as means and counts. Pandas has a native function to do just that, it will output a count, mean, standard deviation, minimum, 25th percentile (Q1), median (50th percentile or Q2), 75th percentile (Q3), and maximum for each numeric columns.
df.describe()| ...1 | ID | Year_Birth | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1754.000 | 1754.00000 | 1735.00 | 1754.000000 | 1754.000000 | 1754.0000 | 1754.00000 | 1754.0000 | 1754.00000 | 1754.00000 | 1754.00000 | 1754.000000 | 1754.000000 | 1754.000000 | 1754.000000 | 1754.000000 | 1754 | 1754 |
| mean | 5584.696 | 1969.57127 | 51166.58 | 0.456100 | 0.480616 | 276.0724 | 28.03478 | 166.4920 | 40.51710 | 28.95838 | 47.26682 | 3.990878 | 2.576967 | 5.714937 | 5.332383 | 0.011403 | 3 | 11 |
| std | 3254.656 | 11.87661 | 26200.42 | 0.537854 | 0.536112 | 314.6047 | 41.34888 | 225.5617 | 57.41299 | 42.83066 | 53.88565 | 2.708278 | 2.848335 | 3.231465 | 2.380183 | 0.106202 | 0 | 0 |
| min | 0.000 | 1893.00000 | 1730.00 | 0.000000 | 0.000000 | 0.0000 | 0.00000 | 0.0000 | 0.00000 | 0.00000 | 0.00000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 3 | 11 |
| 25% | 2802.500 | 1960.00000 | 33574.50 | 0.000000 | 0.000000 | 19.0000 | 2.00000 | 15.0000 | 3.00000 | 2.00000 | 10.00000 | 2.000000 | 0.000000 | 3.000000 | 3.000000 | 0.000000 | 3 | 11 |
| 50% | 5468.000 | 1971.00000 | 49912.00 | 0.000000 | 0.000000 | 160.5000 | 9.00000 | 66.0000 | 13.00000 | 9.00000 | 27.00000 | 3.000000 | 1.000000 | 5.000000 | 6.000000 | 0.000000 | 3 | 11 |
| 75% | 8441.250 | 1978.00000 | 68130.00 | 1.000000 | 1.000000 | 454.0000 | 35.00000 | 232.0000 | 53.50000 | 35.00000 | 63.00000 | 6.000000 | 4.000000 | 8.000000 | 7.000000 | 0.000000 | 3 | 11 |
| max | 11191.000 | 1996.00000 | 666666.00 | 2.000000 | 2.000000 | 1492.0000 | 199.00000 | 1725.0000 | 259.00000 | 263.00000 | 362.00000 | 27.000000 | 28.000000 | 13.000000 | 20.000000 | 1.000000 | 3 | 11 |
8 rows × 18 columns
We can also directly calculate the relevant statistics on numberic columns or rows we are interested in using the functions max(), min(), mean(), median() etc..
df["MntWines"].max()1492df[["Kidhome", "Teenhome"]].mean()Kidhome 0.456100
Teenhome 0.480616
dtype: float64There are also ceartin functions that are usfule for non-numeric columns. To know which stings are present in a object, the fuction unique() can be used, this will returns an array with the unique values in the column or row.
df["Education"].unique()array(['Graduation', 'Master', 'Basic', '2n Cycle'], dtype=object)To know how often a value is present in a column or row, the function value_counts() can be used. This will print a series for all the unique values and print a count.
df["Marital_Status"].value_counts()Marital_Status
Married 672
Together 463
Single 382
Divorced 180
Widow 53
Alone 2
Absurd 2
Name: count, dtype: int648.4.6 Getting summary statistics on grouped data
Pandas is equipped with lots of useful functions which make complicated tasks very easy and fast. One of these functions is .groupby() with the arguments by=..., which will group a DataFrame using a categorical column (for example Education or Marital_Status). This makes it possible to perform operations on a group directly without the need for subsetting. For example, to get a mean income value for the different Education levels in the DataFrame can be done by specifying the column name for the grouping variable by .groupby(by='Education') and specifying the column name to perform this action on [Income] followed by the sum() function.
df.groupby(by="Education")["Income"].mean()Education
2n Cycle 47633.190000
Basic 20306.259259
Graduation 52720.373656
Master 52917.534247
Name: Income, dtype: float68.4.7 Subsetting Questions and Exercises
Here there are several exercises to try conditional subsetting. Try to first before seeing the awnsers.
How many Single people are there in the table that also greduated? And how many are single?
sum(df["Marital_Status"] == "Single")382Subset the DataFrame with people born before 1970 and after 1970 and store both DataFrames
df_before = df[df["Year_Birth"] < 1970]
df_after = df[df["Year_Birth"] >= 1970]How many people are in the two DataFrames?
print("n(before) =", df_before.shape[0])
print("n(after) =", df_after.shape[0])n(before) = 804
n(after) = 950Do the total number of people sum up to the original DataFrame total?
df_before.shape[0] + df_after.shape[0] == df.shape[0]True
print("n(sum) =", df_before.shape[0] + df_after.shape[0])
print("n(expected) =", df.shape[0])n(sum) = 1754
n(expected) = 1754How does the mean income of the two groups differ?
print("income(before) =", df_before["Income"].mean())
print("income(after) =", df_after["Income"].mean())
income(before) = 55513.38113207547
income(after) = 47490.29255319149
Bonus: Can you find something else that differs a lot between the two groups?
This is an open ended question.
8.5 Dealing with missing data
In large tables, it is often important to check if there are columns or rows that have missing data. pandas represents missing data with NA (Not Available). To identify these missing values, pandas provides the .isna() function. This function checks every cell in the DataFrame and returns a DataFrame of the same shape, where each cell contains a Boolean value: True if the original cell contains NA, and False otherwise.
df.isna()| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 1 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 2 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 3 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 4 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1749 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 1750 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 1751 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 1752 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
| 1753 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE |
1754 rows × 20 columns
It is very hard to see if there are any ‘True’ values in this new Boolean table. To investigate how many missing values are present in the table, the sum() function can be used. In Python, True has the 1 assigned to it and False 0.
df.isna().sum()ID 0
Year_Birth 0
Education 0
Marital_Status 0
Income 19
Kidhome 0
Teenhome 0
MntWines 0
MntFruits 0
MntMeatProducts 0
MntFishProducts 0
MntSweetProducts 0
MntGoldProds 0
NumWebPurchases 0
NumCatalogPurchases 0
NumStorePurchases 0
NumWebVisitsMonth 0
Complain 0
dtype: int64In this case is it not clear what a missing value represents, are these individuals who did not want to state their income, or did not have an income? For the tutorial, we are going to keep them in the data.
Two possible actions that can be taken to deal with missing data. One is to remove the row or column using the .dropna() function. This function will by default remove the row with the NA but with specifying the axis=1 the column will be dropped. The second course of action is filling the empty cells with a value, this can be done with the .fillna() function, which will substitute the missing value with a set value, such as 0.
8.6 Combining data
8.6.1 Concatenation exercises
Data is very often present in multiple tables. Think, for example, about a taxonomy table giving count data per sample. One way to combine multiple datasets is through concatenation, which either combines all columns or rows of multiple DataFrames. The function in Pandas that does just that is called .concat. This command combines two DataFrames by appending all rows or columns: .concat([first_dataframe, second_dataframe]).
In the DataFrame, there are individuals with the education levels Graduation, Master, Basic, and 2n Cycle. PhD is missing; however, there is data on people with the education level PhD in another table called phd_data.tsv.
With everything learned so far, and basic information on the .concat()function, try to read in the data from ../phd_data.tsv and concatenate it to the existing df.
Read the tsv “phd_data.tsv” as a new DataFrame and name the variable df2.
df2 = pd.read_csv("../phd_data.tsv", sep="\t")Concatenate the “old” DataFrame df and the new df2 and name the concatenated one concat_df.
concat_df = pd.concat([df, df2])
concat_df| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Z_CostContact | Z_Revenue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 | 3 | 11 |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | 11 |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 | 3 | 11 |
| 3 | 6182 | 1984 | Graduation | Together | 26646 | 1 | 0 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | 0 | 4 | 6 | 0 | 3 | 11 |
| 4 | 7446 | 1967 | Master | Together | 62513 | 0 | 1 | 520 | 42 | 98 | 0 | 42 | 14 | 6 | 4 | 10 | 6 | 0 | 3 | 11 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 481 | 11133 | 1973 | PhD | YOLO | 48432 | 0 | 1 | 322 | 3 | 50 | 4 | 3 | 42 | 7 | 1 | 6 | 8 | 0 | 3 | 11 |
| 482 | 9589 | 1948 | PhD | Widow | 82032 | 0 | 0 | 332 | 194 | 377 | 149 | 125 | 57 | 4 | 6 | 7 | 1 | 0 | 3 | 11 |
| 483 | 4286 | 1970 | PhD | Single | 57642 | 0 | 1 | 580 | 6 | 58 | 8 | 0 | 27 | 7 | 6 | 6 | 4 | 0 | 3 | 11 |
| 484 | 4001 | 1946 | PhD | Together | 64014 | 2 | 1 | 406 | 0 | 30 | 0 | 0 | 8 | 8 | 2 | 5 | 7 | 0 | 3 | 11 |
| 485 | 9405 | 1954 | PhD | Married | 52869 | 1 | 1 | 84 | 3 | 61 | 2 | 1 | 21 | 3 | 1 | 4 | 7 | 0 | 3 | 11 |
2240 rows × 20 columns
Is there anything weird about the new DataFrame and can you fix that?
We previously removed the columns “Z_CostContact” and “Z_Revenue” but they are in the new data again.
We can remove them like before.
concat_df = concat_df.drop("Z_CostContact", axis=1)
concat_df = concat_df.drop("Z_Revenue", axis=1)
concat_df| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 |
| 3 | 6182 | 1984 | Graduation | Together | 26646 | 1 | 0 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | 0 | 4 | 6 | 0 |
| 4 | 7446 | 1967 | Master | Together | 62513 | 0 | 1 | 520 | 42 | 98 | 0 | 42 | 14 | 6 | 4 | 10 | 6 | 0 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 481 | 11133 | 1973 | PhD | YOLO | 48432 | 0 | 1 | 322 | 3 | 50 | 4 | 3 | 42 | 7 | 1 | 6 | 8 | 0 |
| 482 | 9589 | 1948 | PhD | Widow | 82032 | 0 | 0 | 332 | 194 | 377 | 149 | 125 | 57 | 4 | 6 | 7 | 1 | 0 |
| 483 | 4286 | 1970 | PhD | Single | 57642 | 0 | 1 | 580 | 6 | 58 | 8 | 0 | 27 | 7 | 6 | 6 | 4 | 0 |
| 484 | 4001 | 1946 | PhD | Together | 64014 | 2 | 1 | 406 | 0 | 30 | 0 | 0 | 8 | 8 | 2 | 5 | 7 | 0 |
| 485 | 9405 | 1954 | PhD | Married | 52869 | 1 | 1 | 84 | 3 | 61 | 2 | 1 | 21 | 3 | 1 | 4 | 7 | 0 |
2240 rows × 18 columns
Is there something interesting about the marital status of some people that have a PhD?
concat_df[concat_df["Education"]=="PhD"]["Marital_Status"].value_counts()Marital_Status
Married 192
Together 117
Single 98
Divorced 52
Widow 24
YOLO 2
Alone 1
Name: count, dtype: int64There are two people that have “YOLO” as their Marital Status …
8.6.2 Merging
Besides concatenating two DataFrames, there is another powerful function for combining data from multiple sources: .merge(). This function is especially useful when we have different types of related data in separate tables. For example, we might have a taxonomy table with count data per sample and a metadata table in another DataFrame.
The pandas function .merge() allows us to combine these DataFrames based on a common column. This column must exist in both DataFrames and contain similar values.
To illustrate the .merge() function, we will create a new DataFrame and merge it with the existing one. Let’s rank the different education levels from 1 to 5 in a new DataFrame and merge this with the existing DataFrame.
As shown before, there are multiple ways of making a new DataFrame. Here, we first create a dictionary and then use the from_dict() function to transform this into a DataFrame.
The from_dict() function will by default use the keys() of the dictionary as column names. A dictionary is made up of {'key':'value'} pairs. To specify that we want the keys() as rows, the orient= argument has to be set to index. This means that the row names are the dictionary keys().
education_dictionary = {
"Basic": 1,
"2n Cycle": 2,
"Graduation": 3,
"Master": 4,
"PhD": 5
}
education_df = pd.DataFrame.from_dict(education_dictionary, orient="index")
education_df| 0 | |
|---|---|
| Basic | 1 |
| 2n Cycle | 2 |
| Graduation | 3 |
| Master | 4 |
| PhD | 5 |
The resulting DataFrame has the Education level as index “row names” and the column name is 0.
The 0 is not a particular useful name for our new column, so we can use the .rename() function to change this.
education_df = education_df.rename(columns={0: "Level"})
education_df| Level | |
|---|---|
| Basic | 1 |
| 2n Cycle | 2 |
| Graduation | 3 |
| Master | 4 |
| PhD | 5 |
Now that there is a new DataFrame with all the needed information, we can merge it with our previous concat_df on the Education column. The .merge() function requires several arguments: left=, which is the DataFrame that will be on the left side of the merge, in our case concat_df; right=, which is the DataFrame that will be on the right side of the merge, which is education_df; left_on=, specifying the column to merge on from the left DataFrame concat_df, which is Education; and right_index=True, indicating that the right DataFrame education_df should be merged using its index. If the values were in a column instead of the index, we would use right_on= instead.
The right one is education_df and the information is in the index.
merged_df = pd.merge(left=concat_df, right=education_df, left_on="Education", right_index=True)
merged_df| ...1 | ID | Year_Birth | Education | Marital_Status | Income | Kidhome | Teenhome | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumWebPurchases | NumCatalogPurchases | NumStorePurchases | NumWebVisitsMonth | Complain | Level | ...21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5524 | 1957 | Graduation | Single | 58138 | 0 | 0 | 635 | 88 | 546 | 172 | 88 | 88 | 8 | 10 | 4 | 7 | 0 | 3 | NA |
| 1 | 2174 | 1954 | Graduation | Single | 46344 | 1 | 1 | 11 | 1 | 6 | 2 | 1 | 6 | 1 | 1 | 2 | 5 | 0 | 3 | NA |
| 2 | 4141 | 1965 | Graduation | Together | 71613 | 0 | 0 | 426 | 49 | 127 | 111 | 21 | 42 | 8 | 2 | 10 | 4 | 0 | 3 | NA |
| 3 | 6182 | 1984 | Graduation | Together | 26646 | 1 | 0 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | 0 | 4 | 6 | 0 | 3 | NA |
| 5 | 965 | 1971 | Graduation | Divorced | 55635 | 0 | 1 | 235 | 65 | 164 | 50 | 49 | 27 | 7 | 3 | 7 | 6 | 0 | 3 | NA |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | NA |
| 481 | 11133 | 1973 | PhD | YOLO | 48432 | 0 | 1 | 322 | 3 | 50 | 4 | 3 | 42 | 7 | 1 | 6 | 8 | 0 | 5 | NA |
| 482 | 9589 | 1948 | PhD | Widow | 82032 | 0 | 0 | 332 | 194 | 377 | 149 | 125 | 57 | 4 | 6 | 7 | 1 | 0 | 5 | NA |
| 483 | 4286 | 1970 | PhD | Single | 57642 | 0 | 1 | 580 | 6 | 58 | 8 | 0 | 27 | 7 | 6 | 6 | 4 | 0 | 5 | NA |
| 484 | 4001 | 1946 | PhD | Together | 64014 | 2 | 1 | 406 | 0 | 30 | 0 | 0 | 8 | 8 | 2 | 5 | 7 | 0 | 5 | NA |
| 485 | 9405 | 1954 | PhD | Married | 52869 | 1 | 1 | 84 | 3 | 61 | 2 | 1 | 21 | 3 | 1 | 4 | 7 | 0 | 5 | NA |
2240 rows × 19 columns
8.7 Data visualisation
Just looking at DataFrames is nice and useful, but in many cases, it is easier to look at data in graphs. The function that can create plots directly from DataFrames is .plot(). The .plot() function uses the plotting library matplotlib by default in the background. There are other plotting libraries such as Plotnine which will be shown further in the tutorial.
The only arguments .plot() requires are kind=..., and the plot axis x=... and y=.... The kind argument specifies the type of plot, such as hist for histogram, bar for bar plot, and scatter for scatter plot. Check out the Pandas documentation (https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.plot.html) for more plot kinds and useful syntax. There are many aesthetic functions that can help to create beautiful plots. These functions such as .set_xlabel() or ,set_title() are added to the plot, as shown in the examples below.
8.7.1 Histogram
ax = merged_df.plot(kind="hist", y="Income")
ax.set_xlabel("Income")
ax.set_title("Histogram of income")Results in Figure 8.3.
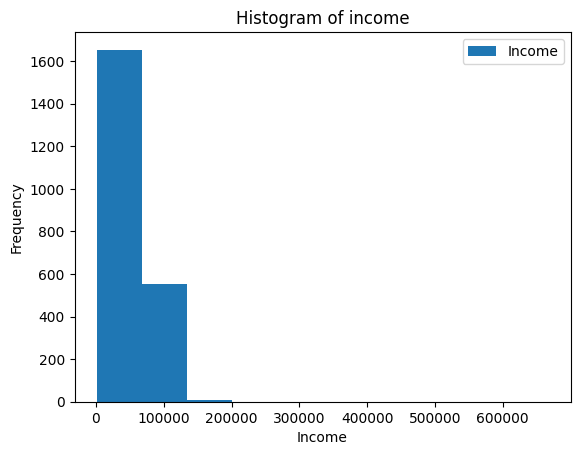
This does not look very good because the x-axis extends so much! Looking at the data, can you figure out what might cause this?
When we look at the highest earners, we see that somebody put 666666 as their income. This is much higher than any other income, which makes the histogram very draged out to include this person.
merged_df[merged_df["Income"].sort_values(ascending=False)]1749 666666.0
1006 157733.0
1290 157146.0
512 153924.0
504 105471.0
...
1615 NaN
1616 NaN
1616 NaN
1621 NaN
1744 NaN
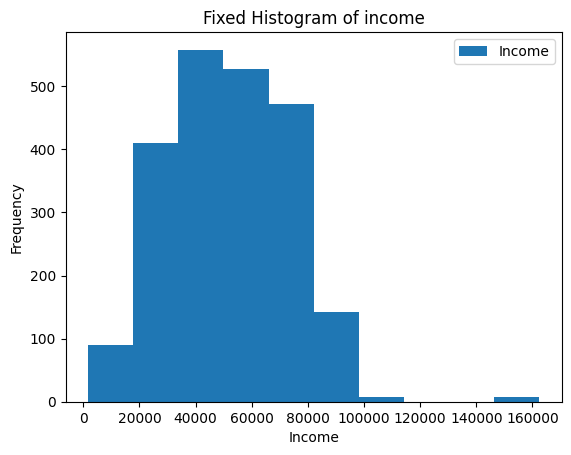
Name : Income, Length: 1754, dtype: float63Use conditional subsetting to make the histogram look nicer.
ax = merged_df[merged_df["Income"] != 666666].plot(kind="hist",y="Income")
ax.set_xlabel("Income")
ax.set_title("Fixed Histogram of income")Results in Figure 8.4.
8.7.2 Bar plot
Instead of making a plot from the original DataFrame we can use the groupby and mean methods to make a plot with summary statistics.
grouped_by_education = merged_df.groupby(by="Education")["Income"].mean()
grouped_by_educationEducation
2n Cycle 47633.190000
Basic 20306.259259
Graduation 52720.373656
Master 52917.534247
PhD 56145.313929
Name: Income, dtype: float64ax = grouped_by_education.plot(kind="bar")
ax.set_ylabel("Mean income")
ax.set_title("Mean income for each education level")Results in Figure 8.5.

8.7.3 Scatter plot
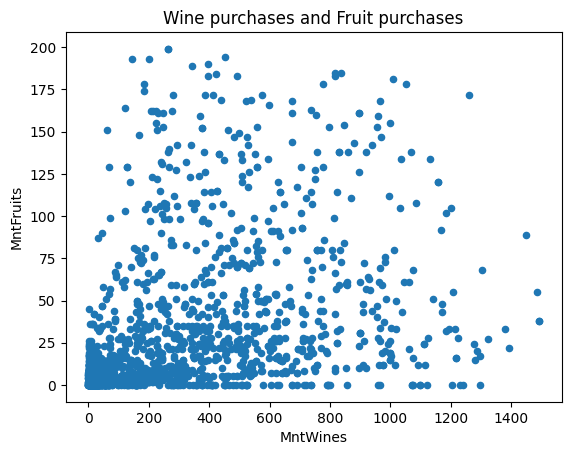
Another kind of plot is the scatter plot, which needs two columns for the x and y axis.
ax = df.plot(kind="scatter", x="MntWines", y="MntFruits")
ax.set_title("Wine purchases and Fruit purchases")Results in Figure 8.6.
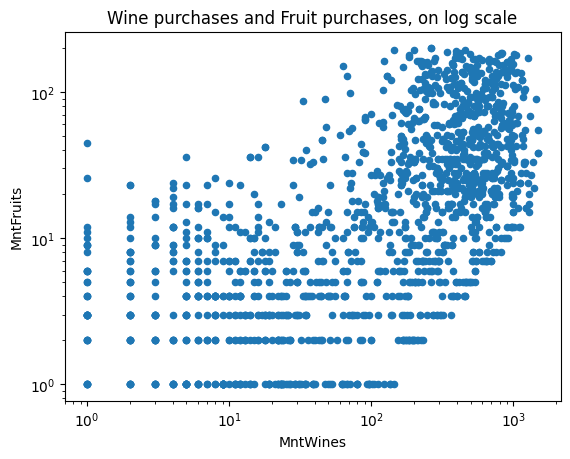
We can also specify whether the axes should be on the log scale or not.
ax = df.plot(kind="scatter", x="MntWines", y="MntFruits", logy=True, logx=True)
ax.set_title("Wine purchases and Fruit purchases, on log scale")Text(0.5, 1.0, ‘Wine purchases and Fruit purchases, on log scale’)
8.8 Plotnine
Plotnine is the Python clone of ggplot2, which is very powerful and is great if we are already familiar with the ggplot2 syntax!
from plotnine import *(ggplot(merged_df, aes("Education", "MntWines", fill="Education"))
+ geom_boxplot(alpha=0.8))Results in Figure 8.7.

(ggplot(merged_df[(merged_df["Year_Birth"]>1900) & (merged_df["Income"]!=666666)],
aes("Year_Birth", "Income", fill="Education"))
+ geom_point(alpha=0.5, stroke=0)
+ facet_wrap("Marital_Status"))Results in Figure 8.8.

8.8.1 Advanced Questions and Exercises
Now that we are familiar with python, pandas, and plotting. There are two data.tables from AncientMetagenomeDir which contains metadata from metagenomes. We should, by using the code in the tutorial be able to explore the datasets and make some fancy plots.
file names:
sample_table_url
library_table_url8.9 Summary
In this chapter, we have started exploring the basics of data analysis using Python with the versatile Pandas library. We wrote Python code and executed it in a Jupyter Notebook, with just a handful of functions such as .read_csv(), .loc[], drop(), merge(), .concat() andplot()`, we have done data manipulation, calculated summary statistics, and plotted the data.
The takeaway messages therefore are:
- Python, Pandas and Jupyter Notebook are relatively easy to use and powerful for data analysis
- If you know R and R markdown, the syntax of Python is easy to learn
These functions in this chapter and the general idea of the Python syntax should help you get started using Python on your data.
8.10 (Optional) clean-up
Let’s clean up your working directory by removing all the data and output from this chapter.
When closing your jupyter notebook(s), say no to saving any additional files.
Press ctrl + c on your terminal, and type y when requested. Once completed, the command below will remove the /<PATH>/<TO>/python-pandas directory **as well as all of its contents*.
Always be VERY careful when using rm -r. Check 3x that the path you are specifying is exactly what you want to delete and nothing more before pressing ENTER!
rm -r /<PATH>/<TO>/python-pandas*Once deleted you can move elsewhere (e.g. cd ~).
We can also get out of the conda environment with
conda deactivateTo delete the conda environment
conda remove --name python-pandas --all -y